Global Microbial Gene Catalog v1.0
Global Microbial Gene Catalog v1.0
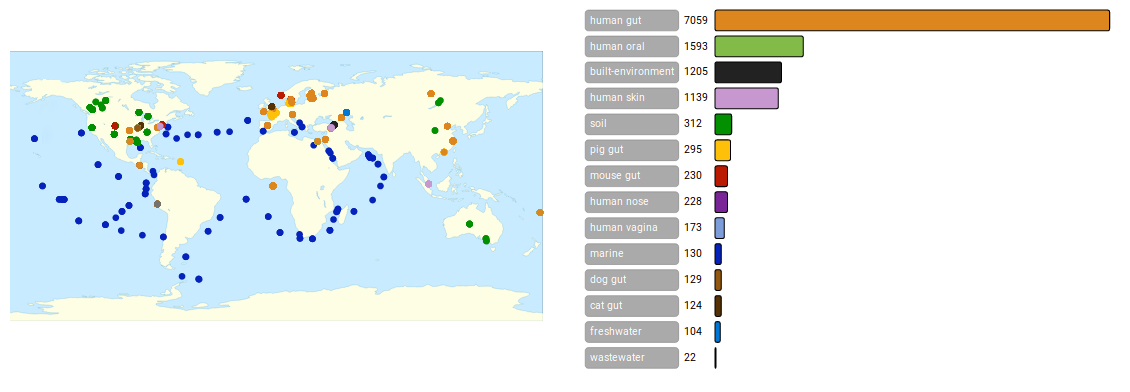
The Global Microbial Gene Catalog is an integrated, consistently-processed, gene catalog of the microbial world, combining metagenomics and high-quality sequenced isolates. A total of 2.3 billion ORFs from 13,174 metagenomes (covering 14 habitats) and the complete ProGenomes2 database were clustered together at 95% nucleotide identity to build a catalog of 302,655,267 unigenes. Read more...
Find homologues by sequence (BLAST-like) or search by identifier
Search for eggNOG orthologous groups
Use the form below to search for eggNOG 5 orthologous groups (OGs) from Bacteria and Archaea levels. Click on any OG in the results list to find the matching unigenes in GMGC. You can search directly for OG identifiers, or their text descriptions.
Mapping a prokaryotic genome to the GMGC
Use the GMGC-mapper tool to identify metagenomic samples where your genome is present, as well as any MAGs (metagenome-assembled genomes) that are similar.
You can download it from Github, where detailed usage instructions are also available.
Mapping a metagenome to the GMGC
There are two possible workflows:
- Using the GMGC as a resource for profiling metagenomes: Use NGLess and its
gmgcmodule. - Assembling and finding novel data: Use NGLess to assemble and predict genes and then use GMGC-mapper to assign those genes to the GMGC.
Global Microbial Genomic Bins collection version 1.0 (GMBC 1.0)
The GMBCv1.0 include >250k genomic bins, grouped into three quality classes: high-quality, medium-quality, and low-quality. High and medium quality bins can be considered MAGs (metagenome-assembled genomes).
Click the taxonomy names to expand / collapse the tree. Click on any number to display the full list of matching bins with that quality class. To display all genome bins for a particular taxonomic class, double click its name.
Search for an antibiotic to display the basic statistics about possible resistance genes in GMGC.